Prioritize labeling of cell imaging samples primarily based on the possible impression of their label on the mannequin’s efficiency.
Adi Nissim, Noam Siegel, Nimrod Berman
One of many predominant obstacles standing in entrance of many machine studying duties is the dearth of labeled knowledge. Labeling knowledge could be exhausting and costly. Thus, many occasions it isn’t cheap to even attempt to remedy an issue utilizing machine studying strategies attributable to lack of labels.
To alleviate this downside, a subject in machine studying has emerged, referred to as Lively Studying. Lively Studying is a technique in machine studying that gives a framework for prioritizing the unlabeled knowledge samples primarily based on the labeled knowledge {that a} mannequin already noticed. We gained’t get into element about it, you possibly can learn Lively Studying in Machine Studying article that deeply explains the fundamentals of it.
Information science in cell imaging is a analysis subject that quickly advances. But, as in different machine learinng domains, labels are costly. To accomodate this downside, we present on this article an finish to finish work on picture cell active-learning classification process of crimson and white blood cells.
Our purpose is to introduce the mix of Biology and Lively Studying and assist others to resolve related and extra sophisticated duties within the Biology area utilizing Lively Studying strategies.
The article is consructed of three predominant elements:
- Cell Picture Preprocessing — the place we present how one can preprocess unsegmented blood cells photos.
- Cell Function Extraction Utilizing Cell Profiler — the place we present how one can extract morphological function from organic cell photo-images to make use of as options for machine studying fashions.
- Utilizing Lively Studying — the place we present an experiment that simulates the usage of energetic studying together with a comparability to not utilizing energetic studying.
The information set
We are going to use a dataset of blood cell photos which was uploaded to GitHub in addition to Kaggle below the MIT license. Every picture is labeled in line with Pink-Blood-Cell (RBC) vs White-Blood-Cell (WBC) courses. There are extra labels for the 4 sorts of WBCs (Eosinophil, Lymphocyte, Monocyte, and Neutrophil) however we didn’t use these labels in our research.
Right here is an instance of a full-sized, uncooked picture from the dataset:
Making a Samples DataFrame
We discovered that the unique dataset accommodates an export.py script which parses the XML annotations right into a CSV desk with the filenames, cell kind labels and bounding containers for every cell.
Sadly, the unique script didn’t embody a cell_id column which is important as a result of we’re classifying particular person cells. So, we barely modified the code to incorporate this column, in addition to select new filenames which embody the image_id and cell_id:
Step one so as to have the ability to work with the info, is to crop the full-sized photos primarily based on the bounding-box coordinates. This leads to funky cell-images:
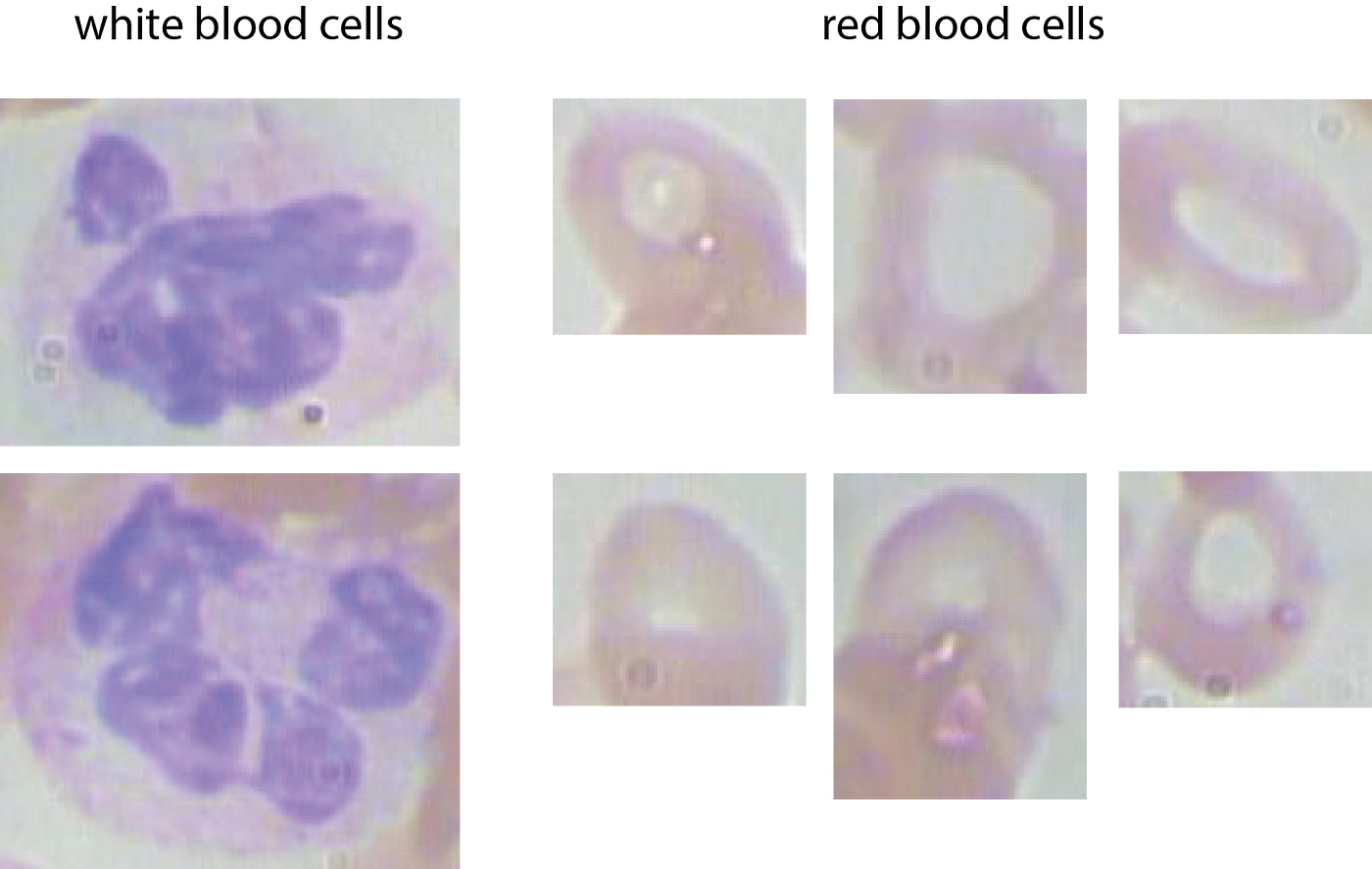
And, right here is the code:
To your comfort, we forked the unique repository and uploaded the cropped photos to GitHub! They are often discovered at:
https://github.com/noamsgl/BCCD_Dataset/tree/grasp/BCCD/cropped
That’s it with the preprocessing! Now, go forward and extract options with CellProfiler.
CellProfiler is a free open-sorce picture evaluation software program, designed to automate quantitative measurement phenotyps from large-scale cell photos. CellProfiler has a user-friendly graphical-user-interface (GUI), permitting you to make use of built-in piplines in addition to constructing your personal tailor-made to your particular wants, utilizing a wide range of measurements and options.
Getting began:
First, obtain Cellprofiler. If CellProfiler is not going to open, chances are you’ll want to put in the Visible C++ Redistributable.
As soon as the GUI pops up, load photos and write organized metadata into CellProfiler. In case you select to construct your personal pipeline, right here chances are you’ll discover the checklist of modules out there via CellProfiler. Many of the modules are divided into three predominant teams: Picture processing, Object processing and Measurements. It can save you your pipeline configuration and reuse it or share it with the world.
Now we are going to reveal the function extraction course of:
- Picture processing – You’ll be able to carry out a number of manipulations on the pictures earlier than activating the options. For instance, changing the picture from shade to grayscale:

- Object processing – These modules detect and establish, or can help you manipulate objects within the picture.

- Measurements – Now, after now we have activated picture processing modules, and now we have recognized major objects in it, measurements identified to be useful for cell profiling may be calculated per object and summarized per picture.

With CellProfiler it can save you the output knowledge to spreedsheets or different sorts of databases. We saved our output as a .CSV file and loaded it to Python as options for the machine studying mannequin.
OK, so now lastly now we have tabular knowledge that we are able to attempt to experiment with. We need to experiment whether or not utilizing an Lively Studying technique can save time by labeling much less knowledge and getting larger accuracies. Our speculation is that utilizing energetic studying, we might probably save valuable effort and time, by lowering considerably the quantity of labeled knowledge required to coach a machine studying mannequin on a cell classification process.
Lively Studying Framework
Earlier than we dive into our experiment, we need to conduct a fast introduction to modAL. ModAL is an energetic studying framework for python. It shares the sklearn API so it’s very easy to combine it into your code if you’re working with Sklearn. This framework lets you simply use completely different state-of-the-art Lively Studying methods. Their documentation is straightforward to comply with and we actually advocate utilizing it in your Lively Studying pipeline.
Lively Studying VS Random appoach
The Experiment
To validate our speculation we are going to conduct an experiment that can evaluate a Random Subsampling technique so as to add new labeled knowledge, to an Lively Studying technique. We are going to begin coaching 2 Logistic Regressions estimators with a number of identical labeled samples. Then, we’re going to use the Random technique for one mannequin and the Lively Studying technique for the second mannequin. We are going to clarify later how that is going to work precisely.
Making ready the Information for the Experiment
We first load the options created by the Cell Profiler. To make issues simpler, we filter platelets that are colorless blood cells. We maintain solely crimson and white blood cells. Thus we try to resolve a binary classification downside – RBC vs WBC. Then we set the label utilizing sklearn label encoder, extract the educational options from the info to X and at last cut up the info to coach and take a look at.
Making ready the info for the experiment
Now we are going to set the fashions
The Dummy learner would be the mannequin that can use the Random technique whereas the energetic learner can be utilizing an Lively Studying technique. To instantiate an energetic learner mannequin, we use ActiveLearner object from modAL bundle. Within the ‘estimator’ subject, you possibly can insert any sklearn mannequin that can be match to the info. The ‘query_strategy’ subject is the place you select a selected Lively Studying technique. We used ‘uncertainty_sampling()’. For extra data try modAL documentation.
Spliting the coaching knowledge
To create our digital methods experiment, we cut up the coaching knowledge into 2 teams. The primary one is coaching knowledge that we all know its labels and we are going to use it to coach our fashions. The second is coaching knowledge that we act like we don’t know its labels. We set the identified practice dimension to five samples.
Simulate Lively VS Dummy
Now, we are going to run 298 epochs the place in every one we are going to practice every mannequin, select the subsequent pattern that we are going to insert the ‘base’ knowledge by the technique of every mannequin, and measure its accuray alongside every epoch. We are going to use common precision rating to measure the preformance of the fashions. We selected this metric due to the imbalanced nature of the dataset.
To select the subsequent pattern within the Random technique, we are going to simply add the subsequent pattern within the ‘new’ group of the dummy dataset. This dataset is shuffled so no want for reshuffling. To select the subsequent pattern with the energetic studying framework, we are going to use the ActiveLearner methodology referred to as ‘question’ which will get the unlabeled knowledge of the ‘new’ group and returns the index of the pattern that he recommends so as to add to the coaching ‘base’ group. Every pattern that we decide will take away from the ‘new’ group in order that no pattern may be choosen twice or extra.
On this instance we added 1 pattern in every iteration. In the actual world, it could be extra sensible to pattern a batch of okay samples at a time.
Outcomes
The variations between the methods are placing!
We will see that utilizing energetic studying we’re capable of attain common precision 0.9 rating with solely 25 samples! Whereas with the dummy technique requires 175 samples to succeed in the identical accuracy! What a niche!
As well as, the Lively mannequin reaches excessive rating of just about 0.99 whereas the dummy stops round 0.95! If we’d use all the info then they’ll attain the identical rating ultimately, however for our objective we restricted to 300 random samples from the dataset.
Code for the plot:
Supervised machine studying methods to cell picture classification require labeled knowledge units. The success of those methods can be constrained by out-of-budget labels as knowledge grows. To beat this problem, we present how one can simply use Lively Studying to pick the fewest variety of photos to label and nonetheless obtain the very best classification scores. Our experiments present that the Lively Studying technique scores 0.9 common precision (a metric appropriate for imbalanced datasets) with simply 25 labels, in comparison with 0.4 with a random choice dummy technique.
Cell imaging has made a big contribution to the fields of biology, medication and pharmacology. It’s used to establish the kind of the cell, its toxicity, integrity and section, and that data is important for healthcare suppliers and scientists to establish illness and broaden our understanding of cell biology.
Beforehand, analyzing cell photos required priceless skilled human capital. Fashionable enchancment within the high quality and availability of microscopy strategies in addition to superior machine studying algorithms, have enabled the info science subject to revolutionize cell imaging. These new applied sciences have led to dynamic and multidimensional high quality databases, and the power to course of this contemporary cell imaging knowledge is having an important position within the development of the cell biology subject.
Hopefully, cell biologists and others will discover this text useful to grasp energetic studying methods, how one can implement them in customized machine studying options, and discover it simpler to make use of knowledge science strategies of their analysis.
Lastly, you might be welcome to share your ideas and options within the feedback part after utilizing Lively Studying to your initiatives!

