To arrange a SolrTextTagger index, or the Aho-Corasick string matching algorithm usually, the essential thought is to populate the FSM with the dictionary entries. Internally, this can create a Trie construction connecting the phrases within the dictionary entries to one another. At runtime, you’d stream textual content towards this construction, and it’ll match phrases within the textual content that match the dictionary entries. The one main drawback of this strategy is that it’ll solely match phrases within the textual content which can be in the identical order because the equal phrase within the dictionary. For instance, if my dictionary entry is “tuberculosis pulmonary”, it is not going to match “pulmonary tuberculosis” within the textual content. Relying in your software, this will or might not be necessary. However I just lately found a strategy to do it, because of the concepts offered within the paper Environment friendly Scientific Idea Extraction in Digital Medical Data (Guo, Kakrania, Baldwin, and Syeda-Mahmood, IBM Analysis Almaden, 2017). Hat tip to my supervisor Ron Daniel for pointing me to the paper.
Particularly, the half that caught my eye was their instance of with the ability to match the string “Chest CT” from the phrase “a subsequent CT scan of the chest”, and their description of their strategy within the paper. As you may see, there’s not a lot element in there, so I’m not certain if my strategy is equivalent to theirs or not, but it surely obtained me pondering, ensuing within the algorithm I’ll describe on this publish. Within the spirit of reproducibility, the outline is accompanied by my Jupyter pocket book containing code to construct the FSM and question it utilizing a memory-based FSM utilizing the PyAhoCorasick library. The pocket book comprises a number of examples of utilizing the Aho-Corasick algorithm to match strings towards an in-house medical dictionary within the inexact method described, towards brief queries (together with the instance cited by the paper), in addition to lengthy paragraph texts.
To hurry up the processing, we suggest a novel indexing technique that considerably reduces the search area whereas nonetheless sustaining the requisite flexibility in matching. First, every phrase within the vocabulary is represented by a novel prefix, the shortest sequence of letters that differentiates it from each different time period. Subsequent, an inverted index is created for the mapping from prefixes to report sentences. Ranging from the consultant prefix of every time period within the vocabulary (or a set of prefixes within the case of a multi-word time period), all related sentences could be simply retrieved as potential matches for the time period, and post-filtering by longest frequent phrase sequence matching can be utilized to additional refine the search outcomes.
The massive thought right here is as follows. As an alternative of populating the FSM with phrases from the dictionary, we populate it with phrases that the phrases are constructed out of. Once we populated it with phrases, the payload related to it was the idea ID. Once we populate it with phrases, the probabilities of collision is sort of sure. For instance, the phrase “tuberculosis” happens 1,352 occasions within the dictionary. So the payload is now a listing of idea ID and synonym ID pairs, very similar to the inverted index construction utilized in engines like google. The idea ID is self-explanatory, the synonym ID is a working serial quantity (for instance, synonym ID 0 is the first identify for the idea). We filter out cease phrases and non-alpha phrases in an try to scale back the noise. Matching is case insensitive (each dictionary and textual content are lowercased) except the phrase is an apparent abbreviation and given in all-caps.
Along with the idea synonym ID pair (CSID hereafter), we additionally retailer within the payload a floating level quantity representing the worth of a match. The concept is that the string “tuberculosis” matched towards the idea for “tuberculosis” is extra useful than a match towards the idea for “pulmonary tuberculosis” or “pulmonary tuberculosis signs”. The burden for every time period within the synonym is the reciprocal of the variety of phrases (minus stopwords) within the synonym. This shall be used later to grade the standard of the matches. Thus the payload for every time period is a listing of pairs (CSID, weight) for every prevalence of the time period in a synonym.
As a result of the PyAhoCorasick API would not permit us to incrementally add to the payload, we preprocess the TSV file representing the dictionary to create a dictionary of phrases and listing of (CSID, weight) pairs. We then use the dictionary to populate an Aho-Corasick FSM. Internally, the FSM states are simply characters, so when querying this construction you’ll have to filter the output, retaining the matches that correspond to the phrases within the question.
When querying, you need to apply the identical set of transformations that you just did on the dictionary phrases, specifically lowercasing except an apparent abbreviation, eradicating stopwords and non-alpha phrases. As talked about earlier, you need to filter the outcomes to solely retain matches to full phrases. The subsequent step is to post-process the outcomes to extract a set of inexact matches.
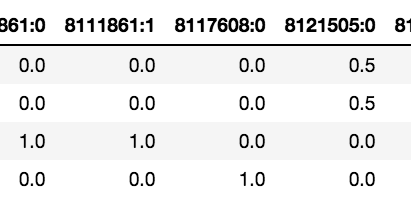
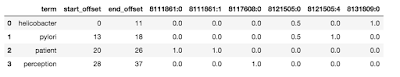
Step one is populate a weight matrix W utilizing the weights from the payload of the filtered outcomes. The rows of this matrix corresponds to the variety of phrases being matched, and the columns correspond to the variety of CSID entries throughout all of the phrases. As a result of phrases would are likely to clump round CSIDs, the matrix is anticipated to be comparatively sparse. The standard of a match of your entire question string towards a selected CSID is evaluated because the sum of the weights down the corresponding column. You present a confidence threshold which the algorithm will use to retain solely matches whose row sum exceeds it. These are actually moved to the candidate match matrix C. The determine under reveals the offsets and weights returned for various CSIDs for the question “helicobacter pylori affected person notion”.
Some attention-grabbing issues to notice right here. Discover that each “helicobacter” and “pylori” have their very own matches (8131809:0 and 8121505:4). As well as, each matched CSID 8121505:0. The time period “affected person” matches two CSIDs precisely — 811861:0 and 811861:1. It’s because the time period is repeated throughout the synonyms, one with a capital P and one with out. Nonetheless, there could also be circumstances the place the identical time period could map to a number of completely different idea IDs as a result of they’re genuinely ambiguous.
The subsequent step is to merge offsets. That is the place we take phrases that are consecutive or shut to one another and contemplate them a single span. Closeness is outlined by a person specified span_slop which is ready to five by default. In our instance above, the primary two phrases could be merged for CSID 8121505:0. At this level, we’ve moved from single phrases annotated to a number of CSID pairs, to some a number of phrases annotated as effectively.
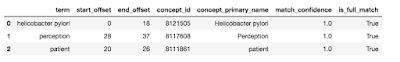
We now take away the mappings that are wholly contained inside longer mappings. Right here the thought is {that a} string corresponding to “helicobacter pylori” is extra particular than both “helicobacter” or “pylori”, and the latter mappings needs to be eliminated in favor of the previous. So that is what the algorithm does subsequent. We are actually prepared to drag within the main identify of the matched idea and current the ultimate outcomes, as proven under.
And that is just about it. This method is relevant not solely to brief strings such because the one I used for the instance, but in addition longer our bodies of textual content. In my Pocket book with the Code and Examples, I present the identical algorithm getting used to map total paragraphs, though it could be higher for high quality causes to annotate sentence by sentence.
In some unspecified time in the future, I hope to fold this performance in to SoDA, my open supply wrapper to SolrTextTagger I discussed earlier. However given my present queue, I do not foresee that taking place anytime quickly. If any of you wish to implement this concept in Scala and combine it into SoDA, please ship me a pull request, and I shall be pleased to merge it in and provide you with credit score.